NLP in Healthcare: Entity Linking
Healthcare and life sciences industry generates a large amount of data, which is driven by compliance & regulatory requirements, record keeping, research papers, etc. But also, with an increase in the amount of data, the search for the necessary documents and articles for research purposes as well as the structuring of data, becomes a more complicated and time-consuming procedure. For example, nowadays, biomedical search engines contain more than 40 million articles, which represent a huge amount of useful healthcare information. But manual processing of such an amount of data is an impassable path, and tools like Natural Language Processing (NLP) and Entity Linking above all facilitate information extraction from text data.
NLP tools, one branch of AI, include applications such as speech recognition, text analysis, translation, and other goals related to language. NLP offers the ability to extract valuable insights from semi-structured/unstructured Clinical and Emerging Data by reading and understanding the data, then translating it into understandable structured data. The idea behind NLP is to connect humans with the processing power of computers better to enhance care and quicken the delivery of treatments, and accelerate research.
Identification of medical terms in free text is the first step in NLP tasks as automatic indexing of biomedical literature and extraction of patients’ problem lists from the text of clinical notes. There are many medical terms that can be found in biomedical literature: diseases names (tuberculosis, glioma, diabetes), symptoms (acute headache, fever, abdominal pain), treatments (chemotherapy, drug therapy), diagnostic tests (biopsies, optical coherence tomography, electrocardiogram), chemicals, anatomical structures, and etc. Along with the detection of medical terms in the text, each of them should be linked to the medical coding standard: International Statistical Classification of Diseases and Related Health Problems (ICD-11), Unified Medical Language System (UMLS), Current Procedural Terminology (CPT) and many other. Particularly, to successfully utilize the plenty of knowledge contained in biomedical records, it is critical to have automated indexing techniques. One of the concepts in the NLP area is called Entity Linking, which helps to solve this task.
Entity linking for a concept detection
In the healthcare domain, accurate entity linking is crucial to correctly understanding the biomedical context. While working with biomedical concepts, one may stumble upon a number of problems: many distinct entities can have very similar mentions, entities may be mentioned in texts through a variety of spelling forms, and entities in the form of abbreviations may be not expanded in a unique way. With these challenges, failure in entity linking (EL) will lead to incorrect interpretation of the contextual information. In the healthcare domain, such faults may lead to risks in medical-related decision-making.
An additional feature of the EL in the healthcare domain is the limited availability of publicly available biomedical EL datasets. At least, it complicates the process of constructing and training entity linking models; assuming the amount of inference data, such EL models may be not generalizable enough. But, in the worst case, some classes of biomedical entities may be not registered in publicly available datasets, which leads to the manual construction of such training sets for a given class of entities.
That is why EL on biomedical texts differs in many ways from that texts from other domains. And thus, solving such challenging tasks requires sophisticated approaches.
Data and methods
Glossary and notations
Entity – named word or phrase (names of diseases, genes, drugs, etc.). Usually, an entity is drawn from the knowledge base.
Knowledge base – a dictionary of entities; usually includes canonical names, definitions, synonyms, etc.
Mention (of entity) – the names of entities in text. Also, contextual (surrounding) information may be considered as a mention.
Entity Linking – mapping mention of an entity from the text to its identity in the knowledge base.
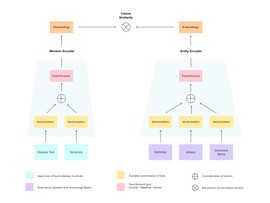
Model Overview
As the EL model, we consider the Siamese neural network, which aims at learning similarities between mentions of entities and corresponding concepts.
We build the following model (see Figure 1). Two branches of the neural network correspond to the mention input and entity input accordingly. Each branch maps the entire text into dense vectors (text of mentioned entity and sentence with this mention from one side and entity-level information from the knowledge base from another). During the training, the model learns to increase cosine similarity between vectors drawn from the correctly linked pair mention+entity and decrease it for incorrectly paired vectors. At the inference stage, each input mention is mapped into the vector space, and the nearest entity vector is assumed to be a vector of the corresponding entity.
However, this approach has several disadvantages, especially with biomedical data.
Issues in biomedical Entity Linking
In the presence of a few biomedical EL datasets only, preparing an accurate training dataset is an important task. For example, considering the disease concept linking task, one can collect information for the training from the following datasets:
- NCBI-disease dataset. The public release of the NCBI disease corpus contains 6.9k disease mentions, which are mapped to 0.8k unique disease concepts (from MeSH and OMIM ontologies). The internal precision of the NCBI-disease dataset is <90%.
- MedMentions is a corpus of Biomedical papers annotated with mentions of UMLS entities. It comprises 4392 abstracts with 34k unique UMLS IDs. This dataset is not a set of diseases only. The internal precision of MedMentions is 97.3%. With keeping 22 UMLS types, which cover diseases and biological processes and entities which have definitions, one could obtain a set of 4805 UMLS disease-like entities. These diseases are labeled in almost all abstracts and cover 15k mentions of 4.8k unique diseases.
- BC5CDR corpus consists of 1.5k PubMed articles with annotated chemicals, diseases, and chemical-disease interactions. By removing chemicals and keeping the diseases only, one could obtain a set of 1.5k abstracts, which cover 3.1k mentions of diseases. These mentions could be mapped into the 1k unique MeSH entities.
As one can see, the total number of the collected abstracts can reach 7k, while the number of mentions/entities in it – about 30k/5k, respectively. This indicates that our training data is highly variegated by the mentions of entities. Also, the vast majority of training samples comprise redundant information (words in abstracts that have nothing in common with diseases).
But the considered model maps the indices of words into the embeddings, and learning such mapping with very large dictionaries with little useful information may be not sufficient and effective. This problem may be tackled in the following ways:
- Using pre-trained embeddings (trained embeddings of biomedical words, e.g. BioWordVec)
- Filtering the dictionary and keeping the useful information only (e.g. words, which are particles of mention phrases, words from knowledge bases)
With the second approach, one may squeeze the size of the dictionary in 10-100 times, which would be beneficial for training and inference speed-up.
Augmentations
Putting the contextual information with the mention into the mention branch of the model may increase the quality of entity linking. Some mentions may be not linked correctly with the mention phrase only, and context could play a key role during the linking. With this, the model can accept, expecting the mention, the sentence in which the mention is cited.
This approach may be developed to increase the generalization ability of the model due to the opening possibility to augment the mentions and corresponding contexts. For example, having a pair of “mention + sentence from abstract with a mention” and “entity from a knowledge base”, one could:
-create negative (non-linked) pairs, using the random words from a sentence instead of the mention;
-expand the mentions by the forward and/or backward words from the sentence;
-remove random parts of sentences without a mention parts.
Training procedure
Since the task is metric learning, the model has to be trained to find similarities between correct “mention” – ”entity” pairs and dissimilarities between incorrect pairs. Such a model can’t be trained on the positive pairs only because it will not be trained to differentiate between correct and incorrect links. On the other side, we can’t assign for each mention all negative entities because our dataset will be expanded by the power-law with the number of mentions, which may be challenging for the relatively big training EL datasets.
So, to train the entity linking model, some sampling procedure has to be considered, for example, triplet loss, which works with the triplet “mention” – ”correct entity” – ”incorrect entity”. Also, one may use special in-batch sampling: the batch of positive pairs could be expanded by negative samples by assigning for each mention in the batch a random entity from a given batch linked to another mention.
Some boosting may be obtained via handling hard negative samples, i.e. incorrectly linked entities, which are more similar for a given mention than the corresponding entity.
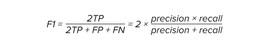
Quality metrics
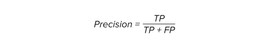
EL quality was measured for each individual sentence by comparing the true and predicted IDs of the concept names with specified cosine similarity cut-off. The method for assessing the quality of entity linking of concept names is in terms of True Positive (TP), True Negative (TN), and False Negative (FN).
Example:
- True disease labeling (upper ID is true): Widespread cognitive disordersD003072, such as deliriumD003693 or ADD010302, have not been previously linked with high blood levels of SSRIs.
- Predicted diseases (upper ID is true, bottom ID is predicted): Widespread cognitive disordersD003072D003072, such as deliriumD003693 or ADD010302D0220454, have not been previously linked with high blood levels of SSRIsD001658.
- TP: cognitive disordersD003072D003072 – when ID_true is equal to ID_pred.
- FP: SSRIsD001658 , ADD010302D0220454 – when ID_pred is not equal to ID_true.
- FN: deliriumD003693 – when ID_true is not equal to ID_pred.
As the accuracy metrics, we used several metrics for better model accuracy measurement:
–Matthews correlation coefficient
Results and Conclusions
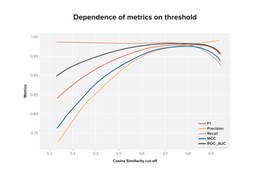
In a resulting model, we obtained the next scores on the validation dataset (see Figure 2).
We discovered that the best quality of separation of correct and incorrect descriptions for a concept is observed at a similarity of around 0.8. That is if the cosine similarity between the description and concept vectors is greater than 0.8, then most likely, this pair is correct.
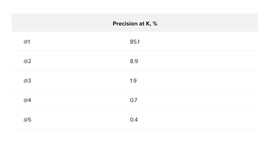
And for the calculation of Precision at K, it is necessary for each concept from the validation sample to select the K nearest descriptions and remember the number of the correct description. If the model were perfect, then for each concept, the closest (K = 1) description would be correct. We performed this experiment on the original (non-multiplied) validation sample. As a result, we got the following values for Precision at K = 5 (see Table 1).
Precision at K showed that for concepts from the validation sample, the closest description would be correct with an accuracy of 85%, and the probability that the correct description will be in the top 5 nearest is 97%.
Also, read our case study in determining how gene mutations affect diseases using NLP.